[Science news] - The flowering plants tree of life

An estimated 350,000 species of flowering plants (angiosperms) are currently known. All are related through common ancestors, and their relationships can be visualised in a phylogenetic tree (similar to a human family tree). Most flowering plant families have had their phylogenies investigated independently. However, there are very few large-scale evolutionary trees that consider all flowering plants.
In a joint study with scientists from Belgium, France, South Africa, the UK, and the Netherlands, we were able to construct a large-scale phylogeny using 36,101 species – nearly one-eighth of all flowering plants. This is now the largest dated phylogenetic framework of flowering plants, generated by combining genuine sequence data and fossil calibration points. It covers all known angiosperm orders, 94.5% of all families of flowering plants and 54.6% of all currently accepted genera.
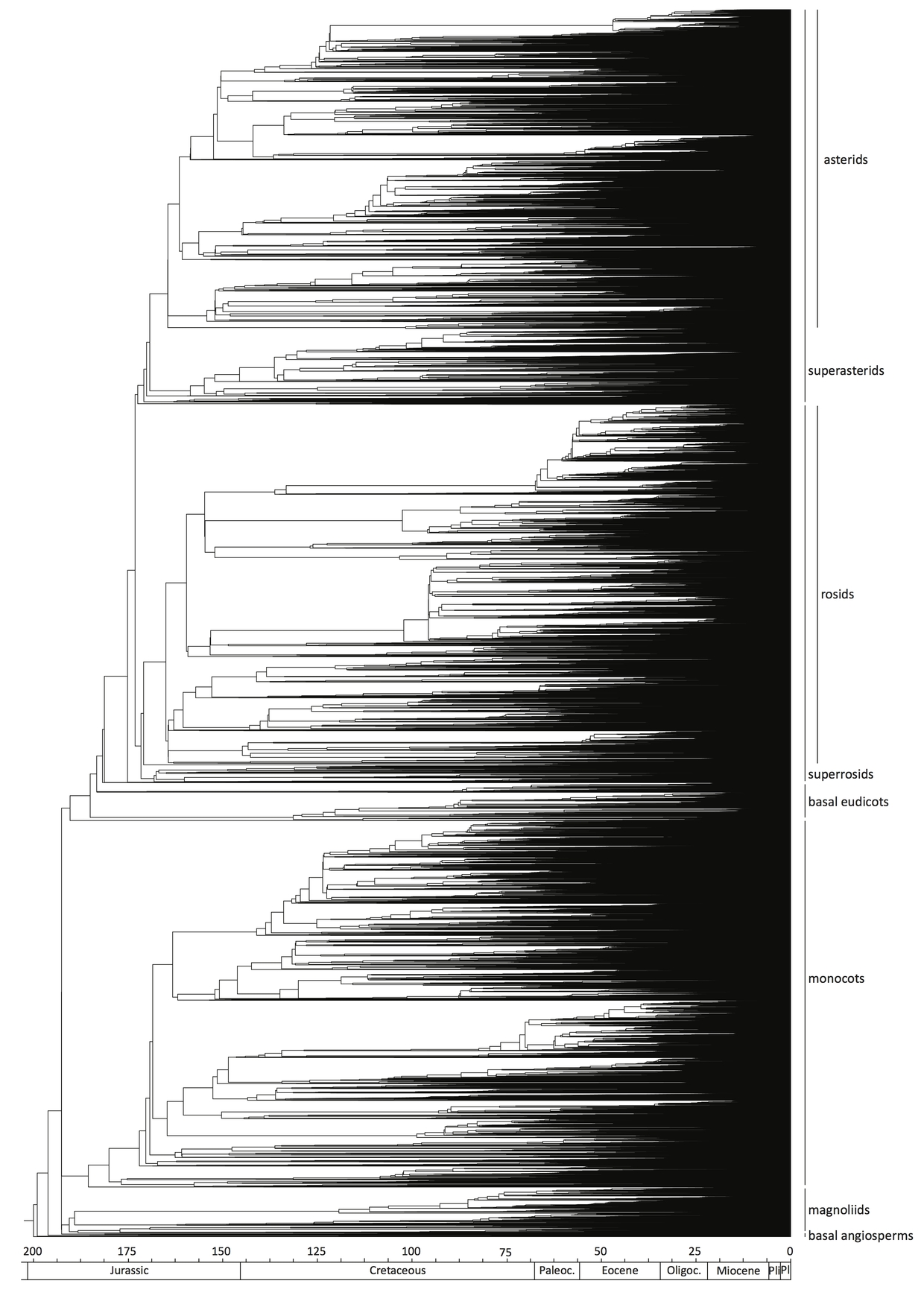 This new evolutionary tree is based on two bar-coding genes on the chloroplast genome. One is highly conserved (good for tracking ‘deep’ ancestors), while the other is very variable (good for detecting recent splits). We also added 56 different fossils to our phylogeny. Fossil plants can be classified at family, genus and sometimes even species level based on their visual characteristics. Fossils of known age, and showing characteristics typical for their group, were used as calibration points. Basically, if fossil evidence shows that a particular group existed at a point in time, we could use it to set our DNA ‘clocks’.
This new evolutionary tree is based on two bar-coding genes on the chloroplast genome. One is highly conserved (good for tracking ‘deep’ ancestors), while the other is very variable (good for detecting recent splits). We also added 56 different fossils to our phylogeny. Fossil plants can be classified at family, genus and sometimes even species level based on their visual characteristics. Fossils of known age, and showing characteristics typical for their group, were used as calibration points. Basically, if fossil evidence shows that a particular group existed at a point in time, we could use it to set our DNA ‘clocks’.
With such detailed coverage, our new phylogeny will be an indispensable tool for large-scale evolutionary, ecological and biogeographical research.
Relevant publications:
Janssens S, Couvreur TLP, Mertens A, Dauby G, Dagallier L‐P, Abeele SV, Vandelook F, Mascarello M, Beeckman H, Sosef M et al 2019. A large‐scale species level dated angiosperm phylogeny for evolutionary and ecological analyses. Biodiversity Data Journal 8: e39677



